BBO-10203: A First-in-Class Cancer Therapeutic Engineered with AI, Supercomputing, and Structure-Based Precision
Introduction: From Undruggable to Clinically Viable
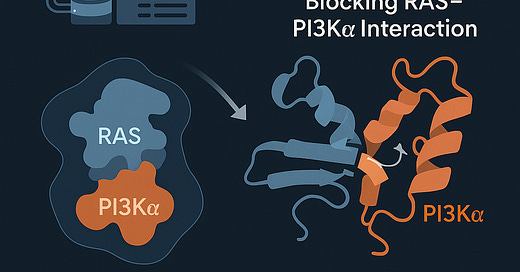
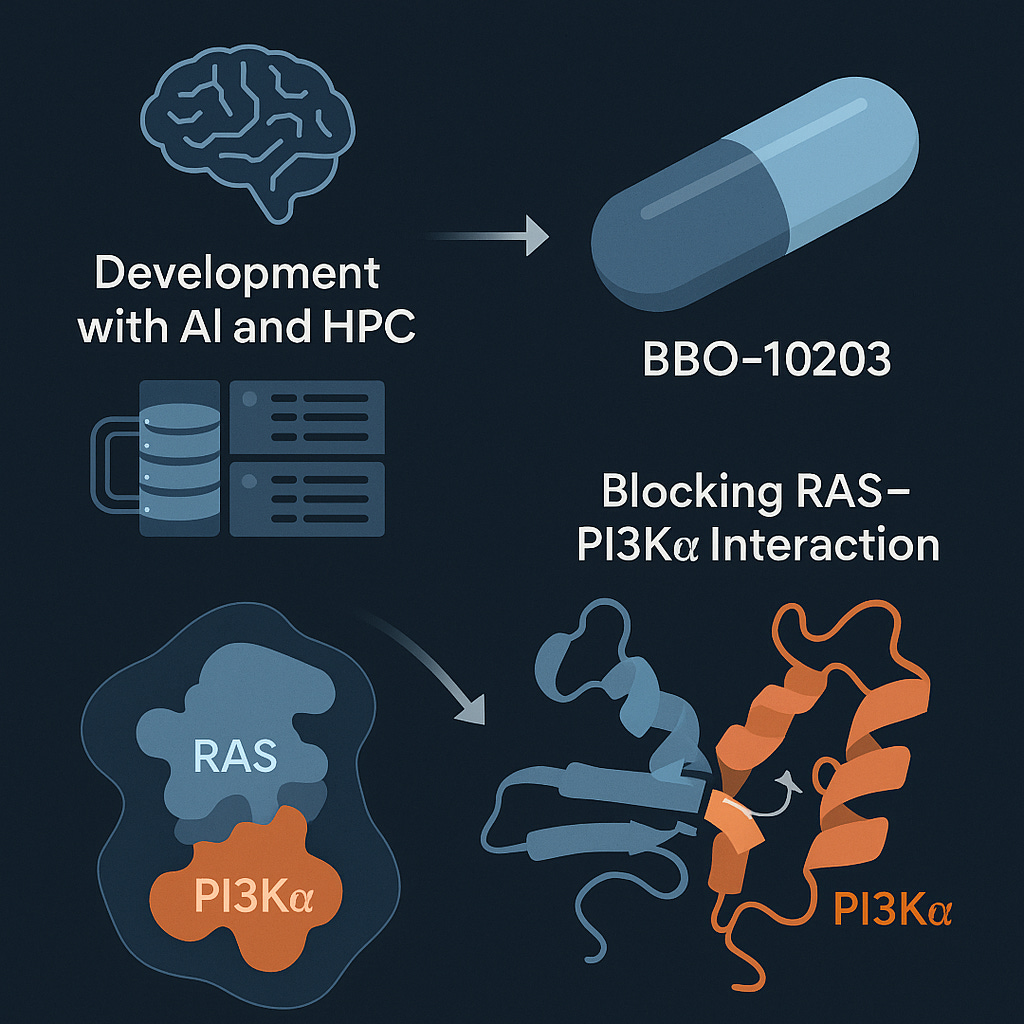
The field of oncology is witnessing a paradigm shift—where once "undruggable" targets like the RAS–PI3Kα interaction posed insurmountable barriers, they are now being cracked open by the fusion of structural biology, AI, and high-performance computing (HPC). A standout example of this convergence is BBO-10203, a first-in-class small molecule that selectively blocks the RAS–PI3Kα protein–protein interaction (PPI), a major cancer-driving mechanism.
Developed by a collaborative effort between Lawrence Livermore National Laboratory (LLNL), BridgeBio Oncology Therapeutics (BBOT), and the Frederick National Laboratory for Cancer Research (FNLCR), BBO-10203 is now in Phase 1 clinical trials—and it’s not just a promising molecule, it’s a new model for how modern drugs should be built.
The Scientific Challenge: Targeting the RAS–PI3Kα Interaction
The RAS and PI3Kα proteins are critical signal transducers in cell growth and survival pathways. Their interactions are implicated in a wide array of aggressive cancers—yet traditional drug discovery approaches have consistently failed to disrupt this interface effectively without adverse side effects, such as hyperglycemia caused by off-target effects on insulin signaling.
This PPI has long stood as a notoriously difficult target in the drug discovery world—until now.
The Breakthrough: From ‘Molecular Glue’ to ‘Molecular Breaker’
Initial structural biology work at FNLCR focused on stabilizing the RAS–PI3Kα interaction using a “molecular glue.” But by analyzing more than 50 high-resolution protein structures, the team flipped the paradigm: designing a compound that could break the interface instead of reinforcing it.
BBO-10203 emerged from this strategy. It selectively binds and disrupts the RAS–PI3Kα interface—without interfering with insulin pathways—leading to a reduction in tumor growth in HER2+, KRAS-mutant, and PIK3CA-mutant cancers.
How It Was Built: HPC-Driven Drug Discovery at Scale
At the core of this innovation lies the Livermore Computer-Aided Drug Design (LCADD) platform at LLNL. LCADD integrates:
AI/ML algorithms
Physics-based molecular simulations
Supercomputing capabilities via Ruby and Lassen (DOE resources)
The Design Loop:
Input: A library of early “glue” compounds and structural insights from crystallography
Simulation: Multi-scale modeling to predict how structural modifications affect binding
Refinement: AI-driven prioritization of features to optimize potency, selectivity, and PK
Output: FEP-scored candidates for biochemical and cellular assays
This approach allowed researchers to simulate millions of molecules and narrow them to a few optimal candidates—long before a single synthesis round.
Preclinical Performance and Translational Potential
In vivo and in vitro studies confirmed the compound’s exceptional profile:
Tumor Growth Inhibition in multiple cancer models
High Selectivity for the RAS–PI3Kα interface
No Hyperglycemia—a common side effect in PI3Kα-targeted therapies
Synergistic Potential with existing treatments (e.g., in breast, colorectal, and lung cancers)
BBO-10203 doesn't just work—it works cleanly, making it ideal for combination regimens or first-line targeted therapies.
Clinical Status: Phase 1 Trials and Forward Outlook
As of June 2025, BBO-10203 entered Phase 1 clinical trials involving patients with advanced solid tumors. These include breast, colorectal, and lung cancers frequently driven by RAS mutations.
Objectives:
Evaluate safety
Determine dose-limiting toxicity
Assess preliminary efficacy and PK/PD profile
Early clinical insights will inform combination strategies and future Phase 2 expansion cohorts.
A Computational Blueprint for Future Drug Discovery
This project stands as a blueprint for future drug discovery, demonstrating how computation can collapse timelines and mitigate risks:
Time-to-clinic: ~6 years from concept to IND—a significant reduction vs. 10–15 years for traditional pipelines
Cost efficiency: Fewer wet-lab experiments required due to predictive modeling
Higher hit-rates: Crystallographically validated binding poses before synthesis
The same framework is now being used to develop therapeutics beyond cancer, targeting macrocycles, enzymes, and peptides.
Conclusion: Beyond a Molecule—A New Model for Precision Oncology
BBO-10203 isn’t just another molecule—it’s the embodiment of a computational-first strategy in precision medicine. By uniting structural biology with AI and HPC, LLNL, BBOT, and FNLCR have demonstrated a powerful method to design “undruggable” targets into druggable realities.
As clinical data emerges, BBO-10203 could define a new standard for targeting PPIs, offering a safer and more effective way to fight aggressive cancers.
Medvolt’s Perspective: Advancing FEP-Driven Precision Drug Discovery
At Medvolt, we champion similar principles through our proprietary AI-integrated Free Energy Perturbation (FEP) platform. By combining physics-based simulations, generative modeling, and deep learning, we empower biotech and pharma teams to:
Prioritize compounds with high affinity and selectivity
Reduce experimental screening costs
Predict binding free energies and ligand pathways at atomic resolution
Streamline lead optimization through dynamic simulation and ML-guided ranking
Our approach resonates strongly with the methodologies used in the development of BBO-10203—validating the transformative power of simulation-first strategies in modern drug discovery.
Ready to bring such power to your pipeline?
Explore Medvolt’s FEP Capabilties